Our Science
Neurological Diseases
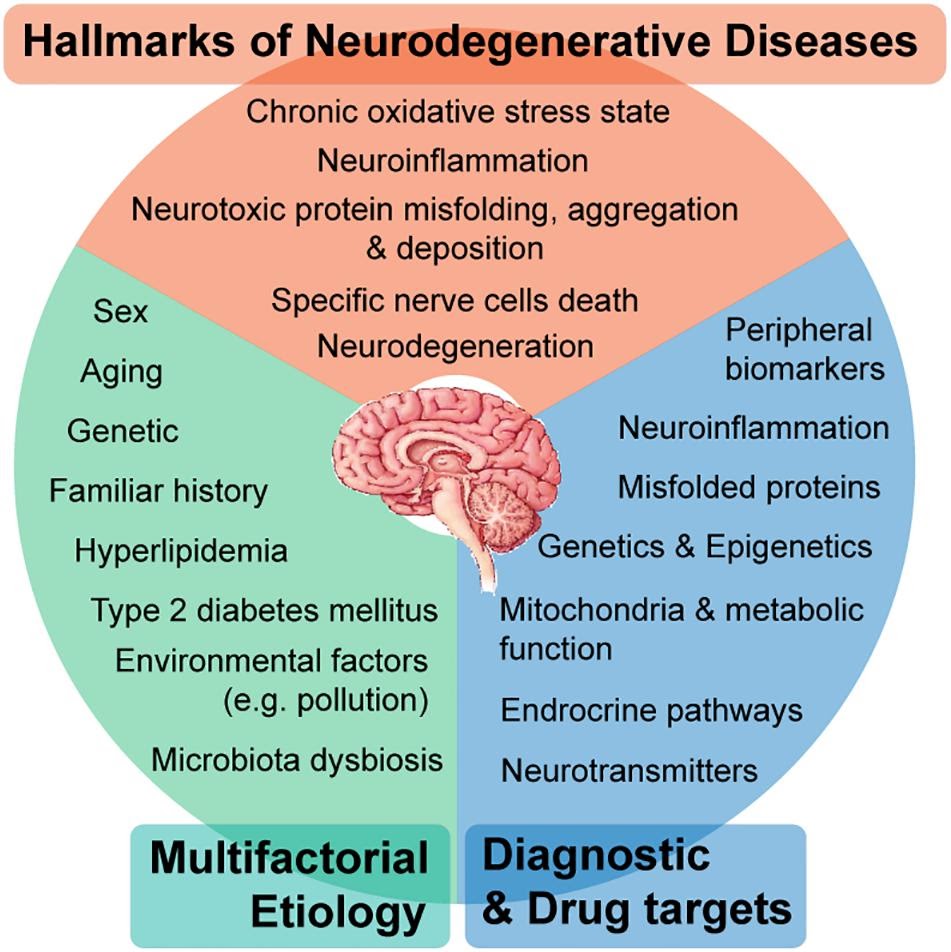
Neurodegenerative diseases (NDDs), such as Alzheimer’s (AD) and Parkinson’s (PD), are among the leading causes of lost years of healthy life and exert a great strain on public healthcare systems. Despite being first described more than a century ago, no effective cure exists for AD or PD. Although extensively characterised at the molecular level, traditional neurodegeneration research remains marred by narrow-sense approaches surrounding amyloid β (Aβ), tau, and α-synuclein (α-syn). NDDs, including AD and PD, are the leading cause of disability-adjusted life years, or lost years of healthy life, accounting for 250.7 million DALYs, or 10.2% of all DALYs, worldwide in 2015. They are also the second-leading cause group of death, with 9.4 million, or 16.8% of global deaths in the same year [1].
The World Health Organization estimates that by 2040 (Gammon, 2014), NDDs will become the second leading cause of death worldwide, taking over cancer and ranking just after cardiovascular diseases (CVD). Global population growth and increased life expectancy mean that ageing-related brain disorders pose an ever-increasing threat in developed countries, placing an unprecedented and growing strain on public healthcare structures. The current therapeutic options for the treatment of AD and PD are limited to simply mitigating neurodegeneration and relieving symptoms rather than reversing damage done. With the advent of high-throughput data generation, integration of data from multiple ‘omics and applying artificial intelligence, we now see evidence to treat AD and PD not as discrete diseases, but as individual, heterogeneous forms of NDD.
Kidney Diseases
Kidney disease, or renal disease, also known as nephropathy, is damage to or disease of a kidney. Nephritis is an inflammatory kidney disease and has several types according to the location of the inflammation. Inflammation can be diagnosed by blood tests. Nephrosis is a non-inflammatory kidney disease. Nephritis and nephrosis can give rise to nephritic syndrome and nephrotic syndrome respectively. Kidney disease usually causes a loss of kidney function to some degree and can result in kidney failure, the complete loss of kidney function. Kidney failure is known as the end-stage of kidney disease, where dialysis or a kidney transplant is the only treatment option.
Chronic kidney disease is defined as prolonged kidney abnormalities (functional and/or structural in nature) that last for more than three weeks. Acute kidney disease is now termed acute kidney injury and is marked by the sudden reduction in kidney function over seven days. About one in eight Americans (as of 2007) have chronic kidney disease, a rate that is increasing over time.
Revealing the Host-Microbiome Interactions

There are functional links between the gut microbiota and its host that may lead to increase in the harvested energy and alterations in the host metabolism. We employ constraint-based modelling of different cell/tissue types in the human body and integrate these models with models for the gut ecosystem. The efforts for modelling of the human metabolism started with the reconstruction of generic human which are literature-based models representing the comprehensive collections of biochemical reactions that occur in the human body. These generic models are useful resources for computational modelling as well as network dependent analysis. However, metabolism varies in each tissue of the human body and it is therefore necessary to reconstruct cell or tissue specific models. In this context, several algorithms (reviewed in Mardinoglu, Nat Rev Gast. 2018) have been developed to generate cell/tissue specific draft GEMs based on generic human models and high-throughput data, e.g., proteomics data from the Human Protein Atlas.
It is feasible to understand more about whole body physiology by studying the interactions between the functional cell/tissue specific models and microbiome together with integration of clinical data (Mardinoglu and Nielsen, 2012). This knowledge can be used to elucidate the progression of metabolic disease, make hypotheses for new therapies and design new clinical interventions. To this end, the intercellular interactions between the metabolically active human cell types including liver, adipose, muscle, heart and kidney may be studied and these models can be used for integration of high-throughput data to find metabolic variations and reaction activities in chronic kidney diseases and kidney cancer. Furthermore, the modelling of the interactions between different organs may provide an increased understanding about the stages of metabolic disorders.
The human microbiome composition can regulate the immune systems, affect the host physiology and disease pathophysiology. Several factors impact the microbiome such as drugs and diet. Understanding the effect of the drug on the host-microbiome interactions can be a key feature for future repositioning. Therefore for an efficient treatment strategies on neurodegenerative diseases the essential role of gut-brain axis has to be considered, as well as in the kidney disease with integration of bacterial bioactive metabolite production into the kidney and whole body metabolism.
Artificial Intelligence

Artificial intelligence (AI) is wide-ranging branch of computer science concerned with building smart machines capable of performing tasks that typically require human intelligence. AI is an interdisciplinary science with multiple approaches, but advancements in machine learning and deep learning are creating a paradigm shift in virtually every sector of the tech industry.
The future of 'standard' medical practice might be here sooner than anticipated, where a patient could see a computer before seeing a doctor. Through advances in AI, it appears possible for the days of misdiagnosis and treating disease symptoms rather than their root cause to move behind us. Think about how many years of blood pressure measurements you have, or how much storage you would need to delete to fit a full 3D image of an organ on your laptop? The accumulating data generated in clinics and stored in electronic medical records through common tests and medical imaging allows for more applications of artificial intelligence and high-performance data-driven medicine. These applications have changed and will continue to change the way both doctors and researchers approach clinical problem-solving.
We used the latest algorithms and methods developed in AI to integrate experimentally observed high throughput data, such as genomes, transcriptomes, proteomes, metabolomes and metagenomes for development of efficient treatment strategies for metabolism related diseases.
Systems Biology
Systems biology is an integrative research strategy that combines experimental and computational biology to identify the molecular mechanisms underlying the components of a complex biological system and to obtain a quantitative description. This rapidly developing field integrates mathematical models with publicly available experimentally observed high throughput data, such as genomes, transcriptomes, proteomes, metabolomes and metagenomes, and reconstructs biochemical networks for Systematic analysis of complex systems using interdisciplinary computational tools. In the context of systems biology, the computational approach focuses on the dynamics and interplay between biological systems such as cells, tissues and organs using a holistic approach rather than reductionism that focuses on individual components and typically excludes information regarding time, space and context
In systems biology, predictive mathematical models are employed for the analysis of given experimental data in a quantitative fashion, for gaining new biological knowledge or for performing predictive simulations. There are two approaches to systems biology: the top-down and the bottom-up approach. The top-down approach is a data-driven process where high-throughput experimental data are analysed with the objective of finding patterns or the function of biological subsystems (or pathways) in the system being studied. By contrast, the bottom up approach is typically hypothesis driven where detailed knowledge of subsystems is reconstructed into a mathematical model that can be used to describe the whole system.
Modelling the whole body biological functions
Understanding the interactions between different tissues and their interactions with the gut microbiota is necessary to gain detailed information about whole- body physiology. To study whole- body biological functions in health and disease states, simulation- ready cell specific or tissue- specific models, including those for the liver, adipose and muscle tissues, intestines, brain and red blood cells, as well as for the gut microbiota, can be integrated. Such knowledge can be used in elucidating the progression of metabolic diseases and for designing new therapies and clinical interventions. To study the crosstalk between liver and other human tissues, the secretion rate of VLDL cholesterol from liver can be measured, and these experimentally derived VLDL cholesterol kinetic data can be integrated with a liver genome- scale metabolic model (GSMM) and other constraints (glucose, ketone bodies, urea, bile acids and lactate) to simulate the dynamics of liver metabolism. The uptake and/or secretion rates of glycerol, fatty acids (FAs) and amino acids from adipose and muscle tissues can also be calculated on the basis of the body composition of each subject and integrated with the liver GSMM. The contribution of the gut microbiota to the host metabolism can also be predicted on the basis of diet, which accounts for the interactions among the microbiota. SCFAs, short- chain FAs.